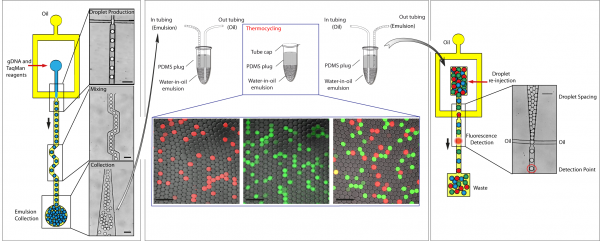
Droplet-based digital PCR microfluidic procedures for genetic and epigenetic alteration(s) detection.
Scientists involved: Gitta Boons, Ouriel Caën, Fanny Garlan, Sonia Garrigou, Heng Lu, Roberta Menezes, Dr Philippe Nizard, Corinne Normand, Dr Karla Perez-Toralla, Dr Shufang Wang-Renault, Dr Eleonora Zonta
Scientists involved previously: Jean-François Bartolo, Zakaria El Harrak, Deniz Pekin, Thevy Hor, Dr Abdel El Abed, Dr Malika Belloul, Raphaël Pantier, Btissem Ziraoui, Dr Yousr Skhiri, Chaouki Ben Salem, Dr. Abdeslam El Harrak, Dr. Linas Mazutis
Genetic and epigenetic alterations within tumor DNA can be used as highly specific biomarkers to distinguish cancer cells from their normal counterparts. These biomarkers are potentially useful for the diagnosis, prognosis, treatment and follow-up of patients. In order to have the required sensitivity and specificity to detect rare tumoral DNA in stool, blood, lymph and other patient samples, a simple, sensitive and quantitative procedure to measure the ratio of mutant to wild-type genes is required. We have developed a procedure allowing the highly sensitive detection of mutated DNA in a quantitative manner within complex mixtures of DNA. In the original work, by using a droplet-based microfluidic system to perform digital PCR in millions of picoliter droplets, we demonstrated the possibility to have an accurate and sensitive quantification of mutated KRAS oncogene in gDNA. This technique enabled the determination of mutant allelic specific imbalance (MASI) in several cancer cell-lines and the precise quantification of mutated KRAS gene in the presence of a 200 000-fold excess of unmutated KRAS genes. The sensitivity is only limited by the number of droplets analyzed. Furthermore, it is also possible to perform multiplex analysis allowing the detection of several different mutations within a single biomarker, mutations in different biomarkers, or even to perform whole targeted pathway analysis. We have now developped droplet-based digital procedures for the highly sensitive detection of a wide range of mutations, copy number variations, DNA hypermethylation and for the quantitative analysis of miRNA and mRNA expression. Future aims of the project are the optimization and validation of this method for its widespread use in clinical oncology.
Fundings
Ligue Nationale Contre le cancer, INCa (Institut National du Cancer), Fondation ARC (Association pour la recherche sur le Cancer), Canceropole Ile de France, CNRS, INSERM, Université Paris-Descartes, Université de Strasbourg, Région Alsace, ANR (French National Research Agency) Nanotechnologies, Aviesan/INSERM (AP physicancer), SIRIC CARPEM.
Institutional Support
CNRS, INSERM, Université Paris-Descartes
Collaborators
Pr Pierre Laurent-Puig (UMRS1147)
Pr Hélène Blons (UMRS1147)
Pr Elisabeth Fabre (UMRS1147)
Dr Nicolas Pecuchet (UMRS1147)
Dr Aziz Zaanan (UMRS1147)
Dr Jean-Baptiste Bachet (UMRS1147)
Dr Anne-Sophie Bats (UMRS1147)
Dr Charlotte Ngo (UMRS1147)
Pr Jean Donadieu, AP-HP, Trousseau Hospital, Paris
Dr Sebastien Héritier, AP-HP, Trousseau Hospital, Paris
Pr Jean-François Emile, AP-HP, Ambroise Paré Hospital, Paris
Pr. Thierry Frebourg, CHU Rouen
Dr Olivier Kosmider, AP-HP, Institut Cochin, Paris
Pr Iradj Sobahni, AP-HP, UPEC, Henri-Mondor Hospital, Paris
Pr Marc Sanson, Brain and Spine Institute (ICM)
Dr Yannick Marie, Brain and Spine Institute (ICM)
Dr J.-C. Baret, Droplets, Membranes and Interfaces Group, Max Plank Institute, Germany
Dr Sylvain Ladame. Biosensor Development Group. Imperial College. London. UK.
Prof. Andrew Griffiths. Laboratory of Chemical Biology. ISIS. Strasbourg. France.
Raindance Technologies. Lexington, MA, US.
PUBLICATIONS
Multiplex detection of rare mutations by picoliter droplet based digital PCR : sensitivity and specificity considerations. E. Zonta, F. Garlan, N. Pecuchet, K. Perez-Toralla, C. Milbury, A. Didelot, O. Caen, H. Blons, E. Fabre, P. Laurent-Puig, V. Taly*. Plos One (2016), 11(7):e0159094 Journal Link.
A Study of Hypermethylated Circulating Tumor DNA as a Universal Colorectal Cancer Biomarker. S. Garrigou, G. Perkins, F. Garlan, C. Normand, A. Didelot, D. Le Corre, S. Peyvandi, C. Mulot, R. Niarra, P. Aucouturier, G. Chatellier, P. Nizard, K. Perez-Toralla, E. Zonta, C. Charpy, A. Pujals, C. Barau, O. Bouché, J.-F. Emile, D. Pezet, F. Bibeau, J. B. Hutchison, D. R. Link, A. Zaanan, P. Laurent-Puig, I. Sobhani and V. Taly*. Clinical Chemistry (2016), 62(8):1129-39. Pubmed.
University Paris Descartes/CNRS/INSERM Press release.
Inserm Press release (english)
Droplet-based digital PCR: application in cancer research. G. Perkins, H. Lu, F. Garlan and V Taly*. Advanced in clinical Chemistry (2016), in press.
Base position error rates analysis of next generation sequencing to detect circulating tumor DNA mutations. N. Pécuchet, Y. Rozenholc, E. Zonta, D. Pietraz, A. Didelot, J-B. Bachet, E. Fabre, V. Taly, H. Blons, P. Laurent-Puig. Clinical Chemistry (2016), in press.
Assessment of DNA integrity, applications for cancer research. Zonta, E., Nizard, P. and Taly, V. Advances in Clinical Chemistry (2015), 70:197-246. doi: 10.1016/bs.acc.2015.03.002. Epub 2015 Apr 11. Review.
Microfluidics and Tumor DNA. Contribution of digital PCR (technical review in French, french title: Microfluidique et ADN tumoral. Apport de la PCR digitale). Nizard, P., Krol, A., Laurent-Puig, P. and Taly, V. Techniques de l'ingénieur (2015). In press.
[Digital PCR compartmentalization I. Single-molecule detection of rare mutations]. Perez-Toralla, K., Pekin, D., Bartolo, J.-F., Garlan, F., Nizard, P., Laurent-Puig, P., Baret, J.-C. and Taly, V. Medecine/Sciences (2015), 31(1):84-92. Review. French. Pubmed.
[Digital PCR compartmentalization II. Contribution for the quantitative detection of circulating tumor DNA]. Caen, O., Nizard, P., Garrigou, S., Perez-Toralla, K., Zonta, E., Laurent-Puig, P. and Taly, V.* Medecine/Sciences (2015), 31(2):180-6. Review. French. Pubmed
Circulating DNA, digital PCR and colorectal cancers. Taly, V and Laurent-Puig, P. Correspondances en Onco-theranostic (2013), 2(4), 184-189 (Invited review for the thematic issue : Cellules tumorales et ADN libre circulant). Journal Link.
Multiplex Picodroplet Digital PCR to Detect KRAS Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients. Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K, Blons H, Bouché O, Landi B, Hutchison JB, Laurent-Puig P. Clin Chem. (2013), In press. Pubmed.
Detecting Biomarkers with microdroplet technology. Taly, V.*, Pekin, D, El Abed A. and Laurent-Puig, P. Invited review. Trends in Molecular Medicine. 2012 Jul;18(7):405-16.
Journal cover Picture.
Quantitative detection of circulating tumor DNA in plasma samples by droplet digital PCR. Pekin D., Kotsopoulos S., Xinyu L., Atochin I., Gang H., Le Corre D., Benhaim L., Hutchison J. B., Link D. R., Blons H., Laurent-Puig P. and Taly V.1,* In: Proceedings of the 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS), 2012.
Quantitative detection of circulating tumor DNA by droplet-based digital PCR. Valerie Taly, Deniz Pekin, Steve Kotsopoulos, Xinyu Li, Ivan Atochin, Hu Gang, Delphine Le Corre, Leonor Benhaim, J. Brian Hutchison, Darren R. Link, Hélène Blons, Pierre Laurent-Puig. American Association for Cancer annual meeting (AACR 2012). March 31-April 4th, Chicago, USA. Cancer Research: April 15, 2012; 72 (8, Sup.1), doi: 10.1158/1538-7445. AM2012-LB-422.
Sensitive detection of rare mutations using droplet-based microfluidics. Baret J.-C. and Taly V. G.I.T. Journal Laboratory Europe (2011), 15: 12-13.
Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Pekin, D., Skhiri, Y., Baret, J.-C., Le Corre, D., Mazutis, L., Ben Salem, C., Millot, F., El Harrak, A., Hutchison, J.B., Larson, J.W., Link, D.R., Laurent-Puig, P., Griffiths, A.D., and Taly, V. Lab on a chip (2011), 11(13): 2156-66.
Highlights in:
Press Release by CNRS, Université de Strasbourg, Université Paris-Descartes and Max Plank Institute (see http://www2.cnrs.fr/presse/communique/2187.htm and followed by Articles in French Press) as well as in the CNRS international Journal. Television (5) and radio (2) interviews.
Journal cover Picture.
Multiplex Digital PCR: Breaking the One Target Per Color Barrier of Quantitative PCR. Zhong, Q., Bhattacharya, S., Kotsopoulos, S., Olson, J., Taly, V., Griffiths, A.D., Link, D.R., and Larson. J.W., Lab on a chip (2011), 11(13):2167-74.
KRAS mutation detection trap. Benhaim, L., Maley, K., Le Corre, D., Blons, H., Taly, V., Bibeau, F., Emile, J-F. and Laurent-Puig, P. Journal of Clinical Oncology (2011), Letter, 29(8): e208-e209.
Multiplex highly sensitive detection of cancer biomarkers in biological samples. D. Pekin, Y. Skhiri, Baret, J.-C., Le Corre, D., Mazutis, L., Ben Salem, C., El Abed, A., Hutchison, J.B., Link, D.R., Griffiths, A.D., Laurent-Puig, P. and Taly, V. In: Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS), 2011. ISBN: 978-0-9798064-3-8.
Droplet-based microfluidics for the quantitative detection of rare mutations. D. Pekin, Y. Skhiri, Baret, J.-C., Le Corre, D., Mazutis, L., Ben Salem, C., El Abed, A., Hutchison, J.B., Link, D.R., Griffiths, A.D., Laurent-Puig, P. and Taly, V. In: Proceedings of the 14th International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS), 2010, p. 58-60. ISBN: 978-0-9798064-3-8.